Python Module obsterms¶
Defining Observables¶
Module obsterms defines the observables on quantum systems. Each observable
contains the corresponding methods to write the fortran files and to read the
results from the fortran outputs. The following observables are defined
Observables on any quantum sytem in OSMPS are specified by instantiating an
object of the obsterms.Observables class, for example
myObservables = Observables(Operators)
One then adds observables using the obsterms.Observables.AddObservable()
method. Each observable is itself a python class and has the corresponding
read/write methods Examples of the use of
obsterms.Observables.AddObservable() are:
siteobservables (obsterms.SiteObservable). Example :
myObservables.AddObservable('site', 'nbtotal', name='n')will measure
at each site and return the result as a length-
vector with the tag
'n'. Recall thatis either the system size in the case of a finite MPS or the unit cell length in the case of iMPS as specified in the table in the description of
WriteFiles()
Site entropy measurement (
obsterms.SiteEntropyObservable).Bond entropy measurement (
obsterms.BondEntropyObservable).corrobservables (obsterms.CorrObservableandobsterms.FCorrObservable). Example:
myObservables.AddObservable('corr', ['nbtotal', 'nbtotal'], name='nn')will measure
at each independent pair of sites
and return the result with the tag
'nn'. For finite-size MPS simulations the result is returned as anmatrix, and for infinite-size MPS simulations the result is returned as a length-
vector, where
is specified by the
SpecifyCorrelationRangemethod, see Sec. Infinite-size ground state search: iMPS. For observables involving odd numbers of fermionic operators on a given site, there is an optionalPhaseargument such as existed for thempo.MPO.AddMPOTerm()method. An example ismyObservables.AddObservable('corr', ['fdagger', 'f'], name='spdm', Phase=True)which measures the fermionic correlation function
.
stringobservables (obsterms.StringCorrObservable). Example (fromInfiniteheisenbergspinone.py):
myObservables.AddObservable('string', ['sz', 'sz', 'phaseString'], name='StringOP')will measure
at each independent pair of sites
and return the result with the tag
'StringOP'. Note thatphaseStringhas been defined to beonly in this particular case.
MPOobservables (obsterms.MPOObservable). Example:
DefectDensity = mps.MPO(Operators) DefectDensity.AddMPOTerm('bond', ['sz', 'sz']) myStaticObservables.AddObservable('MPO', DefectDensity, 'Defect Density')will measure
and return the result with the tag
'Defect Density'. In this way, essentially arbitrary many-body operators may be measured.
Reduced density matrix, limited to one (
obsterms.RhoiObservable) or two (obsterms.RhoijObservable) sites at present, can be measured as well. They can be added once as single item or list. An empty list indicates to report the singular values at all bipartitions. If passed as list, entries have to sorted ascending.
myObservables.AddObservables('DensityMatrix_i', []) myObservables.AddObservables('DensityMatrix_ij', [])
It is possible to measure the mutual information matrix without writing all the information about the reduced density matrices to the result files (
obsterms.MutualInformationObservable).myObservables.AddObservables('MI', True)
The Schmidt values (
obsterms.LambdaObservable) at the bipartitions are another measurement, which gives a more detailed picture than the bond entropy. They can be added once as single item or list. An empty list indicates to report the singular values at all bipartitions.
myObservables.AddObservables('Lambda', [])
Distance to a pure state (
obsterms.DistancePsiObs)Save state independent from checkpoints etc. (MPS only,
obsterms.StateObservable)Gate observables (ED only,
obsterms.GateObservable).Distance to a density matrix (ED only,
obsterms.DistanceRhoObs).
Observables for the different algorithms in OSMPS are specified as part of the
parameters dictionary, see Table in tools.WriteFiles().
Developer’s Guide for new observables
Implement new class in this module, i.e.
obsterms. We need the minimal class functions__init__,add,write, andread.Add new observable to the
dataset andmpslistin :py:class:Observables.Modify
ObsOps_types_templates.f90by adding a new observable toOBS_TYPEand possibly defining a new derived type for this observable.Define read, write, and destroy methods as far as applicable.
Carry out measurements in all
observesubroutines.
- obsterms.check_restore_timeevo(Params)[source]¶
Returns default value.
Arguments
- Paramsdictionary
the dictionary containing the parameters for the simulation.
- class obsterms._ObsTerm[source]¶
- __getitem__(key)[source]¶
Return the corresponding variable from the set of class variabels.
Arguments
- keystring
String corresponding to a class variable.
- default(defargs, kwargs)[source]¶
Set the default arguments for a term if not passed as optional arguments.
- get_hash()[source]¶
Generate hash for this object based on the class dictionary provided as default by python.
- ignored(optargs, kwargs)[source]¶
Print warnings about ignored optional keyword parameters:
Arguments
- optargslist of strings
contains all the string identifiers of the keyword arguments considered.
- kwargsdictionary
contains all the keywords arguments
- line_to_floats(Obsfile, nn)[source]¶
Read line in file handle and convert to 1d numpy array of floats.
Arguments
- Obsfilefile handle
read next line in this file handle
- nnint
number of floats in the line.
- read_rmatrix(fh, d1, d2)[source]¶
Read a matrix in file handle and convert to 2d numpy array of floats.
Arguments
- Obsfilefile handle
read next line in this file handle
- d1int
number of rows in the matrix.
- d2int
number of floats in one line corresponding to the number of columns
- required(args, kwargs)[source]¶
Raise exception for required arguments not present in the optional arguments.
Arguments
- argslist of strings
contains all the string identifiers of the keyword arguments considered.
- kwargsdictionary
contains all the keywords arguments
- setargs(args, argsval, kwargs)[source]¶
Set non key-worded argument in corresponding dictionary.
Arguments
- argslist of strings
contains all the string identifiers of the keyword arguments considered in the correct order.
- argsvallist
contains the values passed by the user as optional non-keyword arguments. Order must be as indicated in documentation.
- kwargsdict
Dictionary with all arguments to be constructed. Here, we set the corresponding non-keyword arguments passed.
- class obsterms.SiteObservable(Ops)[source]¶
Measurement of local observables on each site.
Details
The arguments of the
obsterms.Observables.AddObservable()are- temptypestr
Identification to add site observable. Must be
site.- termstr
Name of the operator to be measured.
- namestr, keyword argument
Key to address measurement in dictionary of results. Default to
<+ term +>.- weightfloat, keyword argument
Weighting the measurement. Default to 1.0
Example
By default the bond entropy is measured. The measurement can be turned off by calling the
obsterms.Observables.AddObservable()function as follows.>>> Obs = mps.Observables(Operators) >>> Obs.AddObservable('site', 'nbtotal')
Variables
The following class variables are defined.
- Oplist of strings
List contains the names of the operators to be measured.
- weightlist of floats
The weights for each measurement.
- namelist of strings
The key under which the results should be stored.
- Opsinstance of
Ops.OperatorList Contains the operators for checks.
- class obsterms.SiteEntropyObservable[source]¶
Measurement of the site entropy of each site along the chain.
Details
The arguments of the
obsterms.Observables.AddObservable()are- termbool
Status of the site entropy measurement’
Truefor measuring,Falsefor not measuring.
Example
By default the site entropy is measured. The measurement can be turned off by calling the
obsterms.Observables.AddObservable()function as follows.>>> Obs = mps.Observables(Operators) >>> Obs.AddObservable('SiteEntropy', False)
Replacing
FalsewithTrue, the site entropy can be turned on.Variables
The following class variables are defined.
- hasbool
Falseif site entropy should not be saved,Trueotherwise measuring the site entropy at every splitting.
- add(term, *args, **kwargs)[source]¶
Add functions as method to turn site entropy measure on and off. Last call determines if the site entropy is measured or not. For arguments see
obsterms.SiteEntropyObservable
- read(fh, **kwargs)[source]¶
Read the results from file. The filehandle must be at the corresponding position.
Arguments
- fhfilehandle
Open file with results. The next lines contain the measurements of the Schmidt decomposition.
- llint, keyword argument
The system size must be given as keyword argument.
- class obsterms.BondEntropyObservable[source]¶
Measurement of the bond entropy at each splitting along the chain.
Details
The arguments of the
obsterms.Observables.AddObservable()are- termtypestr
Identification to modify bond entropy measure. Must be
BondEntropy.- termbool
Status of the bond entropy measurement’
Truefor measuring,Falsefor not measuring.
Example
By default the bond entropy is measured. The measurement can be turned off by calling the
obsterms.Observables.AddObservable()function as follows.>>> Obs = mps.Observables(Operators) >>> Obs.AddObservable('BondEntropy', False)
Replacing
FalsewithTrue, the bond entropy can be turned on.Variables
The following class variables are defined.
- hasbool
Falseif bond entropy should not be saved,Trueotherwise measuring the bond entropy at every splitting.
- add(term, *args, **kwargs)[source]¶
Add functions as method to turn bond entropy measure on and off. Last call determines if the bond entropy is measured or not. For arguments see
obsterms.BondEntropyObservable
- read(fh, **kwargs)[source]¶
Read the results from file. The filehandle must be at the corresponding position.
Arguments
- fhfilehandle
Open file with results. The next lines contain the measurements of the Schmidt decomposition.
- llint, keyword argument
The system size must be given as keyword argument.
- class obsterms.CorrObservable(Ops, FCorr)[source]¶
Measurement of correlation observables on all two-site combinations.
Details
The arguments of the
obsterms.Observables.AddObservable()are- temptypestr
Identification to add correlation observable. Must be
corr.- termlist of two strings or numpy array
Name of the two operators to be measured or numpy 2d array representing an operator on two sites in the Hilbert space to be decomposed.
- namestr, keyword argument
Key to address measurement in dictionary of results. Required.
- weightfloat, keyword argument
Weighting the measurement. Default to 1.0
- Phasebool, optional
If given, must be
False, otherwise Fermi correlation will be measured instead. Default toFalseif not present.
Example
A two site correlation measurement can be added to the observables as follows.
>>> Obs = mps.Observables(Operators) >>> Obs.AddObservable('corr', ['nbtotal', 'nbtotal'], name='<nn>')
Variables
The following class variables are defined.
- Oplist of strings
List contains the names of the operators to be measured.
- intidlist of integers
Index to which correlation pair of operators should be added.
- weightlist of floats
The weights for each measurement.
- namelist of strings
The key under which the results should be stored.
- ishermlist of bools
Checks if observable is hermitian. If not hermitian, the complex part of the correlation is measured as well.
- Opsinstance of
Ops.OperatorList Contains the operators for checks.
- read(fh, **kwargs)[source]¶
Read the results for the correlations from an open file handle at the corresponding position.
- fhfilehandle
Open file with results. The next lines contain the measurements of the correlation matrices.
- read_hdf5(gh5, key, **kwargs)[source]¶
Read the results from an open HDF5 file.
Arguments
- fhid of HDF5 group
This is the HDF5 group containing the observable.
- read_imps(fh, corr_range)[source]¶
Read the results for the correlation for an iMPS simulations. Filehandle must be already open.
- fhfilehandle
Open file handle with results at right position.
- corr_rangeint
Determines number of entries in correlations. For iMPS, the correlations are not a square matrix.
- class obsterms.Corr2NNObservable(Ops)[source]¶
Measure the 4-site correlator between two pairs of nearest neighbors.
Details
The arguments of the
obsterms.Observables.AddObservable()are- temptypestr
Identification to add correlation observable. Must be
corr2nn.- termlist of four strings
Name of the operators acting on the sites i, i+1, j, j+1
- namestr, keyword argument
Key to address measurement in dictionary of results. Required.
- weightfloat, keyword argument
Weighting the measurement. Default to 1.0
Example
This type of four-site correlator of two pairs of nearest neighbors can be added to a measurement as
>>> Obs = mps.Observables(Operators) >>> myObs.AddObservable('corr2nn', ['sigmaz', 'sigmaz', 'sigmaz', 'sigmaz'], name='zzzz')
Variables
The following class variables are defined.
- Oplist of strings
List contains the names of the operators to be measured
- weightlist of floats
The weights for each measurement.
- namelist of strings
The key under which the results should be stored.
- ishermlist of bools
Checks if observable is hermitian. If not hermitian, the complex part of the correlation is measured as well.
- Opsinstance of
Ops.OperatorList Contains the operators for checks.
Details
The resulting matrix is always upper triangular. The measurement of i, i+1, j, j+1 is stored in the matrix element [i, j] and i < j for all measurements. Therefore, the first two columns and the last column is empty. Moreover, the last three rows are empty.
- class obsterms.FCorrObservable(Ops)[source]¶
Measurement of correlation observables on all two-site combinations with a Fermi phase yielded from a Jordan-Wigner transformation.
Details
The arguments of the
obsterms.Observables.AddObservable()are- temptypestr
Identification to add correlation observable. Must be
corr.- termlist of two strings or numpy array
Name of the two operators to be measured or numpy 2d array representing an operator on two sites in the Hilbert space to be decomposed.
- namestr, keyword argument
Key to address measurement in dictionary of results. Required.
- weightfloat, keyword argument
Weighting the measurement. Default to 1.0
- Phasebool, keyword argument
Must be given and must be true for Fermi correlation.
Example
By default the bond entropy is measured. The measurement can be turned off by calling the
obsterms.Observables.AddObservable()function as follows.>>> Obs = mps.Observables(Operators) >>> Obs.AddObservable('corr', ['nftotal', 'nftotal'], name='<nn>', >>> Phase=True)
Variables
The following class variables are defined.
- Oplist of strings
List contains the names of the operators to be measured.
- intidlist of integers
Index to which correlation pair of operators should be added.
- weightlist of floats
The weights for each measurement.
- namelist of strings
The key under which the results should be stored.
- ishermlist of bools
Checks if observable is hermitian. If not hermitian, the complex part of the correlation is measured as well.
- Opsinstance of
Ops.OperatorList Contains the operators for checks.
- read(fh, **kwargs)[source]¶
Read the results for the correlations with Fermi phase from an open file handle at the corresponding position.
- fhfilehandle
Open file with results. The next lines contain the measurements of the correlation matrices.
- read_hdf5(gh5, key, **kwargs)[source]¶
Read the results from an open HDF5 file.
Arguments
- fhid of HDF5 group
This is the HDF5 group containing the observable.
- read_imps(fh, corr_range)[source]¶
Read the results for the Fermi correlation for an iMPS simulations. Filehandle must be already open.
- fhfilehandle
Open file handle with results at right position.
- corr_rangeint
Determines number of entries in correlations. For iMPS, the correlations are not a square matrix.
- class obsterms.StringCorrObservable(Ops)[source]¶
Measurement of a string correlation observables on all two-site combinations with an additional operator applied to sites in between, e.g., a Fermi phase yielded from a Jordan-Wigner transformation.
Details
The arguments of the
obsterms.Observables.AddObservable()are- temptypestr
Identification to add correlation observable. Must be
corr.- termlist of three strings
Name of the two operators to be measured. The third operator is the phase operator.
- namestr, keyword argument
Key to address measurement in dictionary of results. Required.
- weightfloat, keyword argument
Weighting the measurement. Default to 1.0
Example
You can add a string correlation observable by calling the
obsterms.Observables.AddObservable()function as follows.>>> Obs = mps.Observables(Operators) >>> Obs.AddObservable('string', ['sz', 'sz', 'phasestring'], >>> name='StringOP', Phase=True)
Variables
The following class variables are defined.
- Oplist of strings
List contains the names of the operators to be measured. It is a listed list.
- weightlist of floats
The weights for each measurement.
- namelist of strings
The key under which the results should be stored.
- Opsinstance of
Ops.OperatorList Contains the operators for checks.
- read(fh, **kwargs)[source]¶
Read the results for the correlations with Fermi phase from an open file handle at the corresponding position.
- fhfilehandle
Open file with results. The next lines contain the measurements of the correlation matrices.
- read_hdf5(gh5, key, **kwargs)[source]¶
Read the results from an open HDF5 file.
Arguments
- fhid of HDF5 group
This is the HDF5 group containing the observable.
- read_imps(fh, corr_range)[source]¶
Read the results for the string correlation for an iMPS simulations. Filehandle must be already open.
- fhfilehandle
Open file handle with results at right position.
- corr_rangeint
Determines number of entries in correlations. For iMPS, the correlations are not a square matrix.
- class obsterms.MPOObservable[source]¶
Measurement of an MPO beyond the default measurement of the Hamiltonian MPO.
Details
The arguments of the
obsterms.Observables.AddObservable()are- termtypestr
Identification as MPO measurement, string must be ‘MPO’.
- terminstance of class
MPO.MPO. The MPO to be measured.
- namestr, keyword arguments
The key to access the result in the result dictionary. Must be present.
The MPO observables should not depend on time-dependent Hamiltonian parameters. They are not constructed for each time step.
Example
You can add an MPO observable by calling the
obsterms.Observables.AddObservable()function as follows:>>> DefectDensity = mps.MPO(Operators) >>> DefectDensity.AddMPOTerm('bond', ['sz', 'sz']) >>> >>> Obs = mps.Observables(Operators) >>> Obs.AddObservable('MPO', DefectDensity, name='DefectDensity')
Variables
- Oplist of
MPO.MPO Contains the MPO for each measurement in the list.
- namelist of strings
The key under which the results should be stored.
- weightlist of floats
The weights for each measurement.
- class obsterms.RhoiObservable[source]¶
Defines the measurement of single site reduced density matrices.
Details
The arguments of the
obsterms.Observables.AddObservable()are- termtypestr
Identification as single site reduced density matrig, string must be ‘DensityMatrix_i’.
- termlist of integers
Indices of the sites to be measured. If list is empty, it is filled with all sites.
Example
>>> Obs = mps.Observables(Operators) >>> Obs.AddObservables('DensityMatrix_i', [])
Variables
- has_rhoibool
Flag if any density matrix is measured
- ij
None, list of integers Contains the indices of the reduced density matrices to be measured.
- ij_orig
None, list of integers Contains the original list past by the user; this needs to be stored since empty list in ‘ij’ are overwritten with the sites 1 .. L.
- read(fh, **kwargs)[source]¶
Read the local density matrices.
Arguments
- fhfilehandle
Open file with results. The next lines contain the measurements of the correlation matrices.
- paramdictionary (as keyword argument)
Contains the settings for the simulation.
- dynamicbool (as keyword argument)
Flag if dynamics (True) or statics (False) are read.
- ldint (as keyword argument)
Local dimension of the Hilbert space.
- class obsterms.RhoijObservable[source]¶
Defines the measurement of two-site site reduced density matrices.
Details
The arguments of the
obsterms.Observables.AddObservable()are- termtypestr
Identification as two-site reduced density matrix, string must be ‘DensityMatrix_ij’.
- termlist of integers
Pairs of Indices of the sites to be measured. If list is empty, it is filled with all sites. Indices must be ascending.
Example
>>> Obs = mps.Observables(Operators) >>> Obs.AddObservables('DensityMatrix_ij', [])
Variables
- has_rhoijbool
Flag if any density matrix is measured
- ij
None, list of integers Contains the pairs of indices of the reduced density matrices to be measured.
- ij_orig
None, list of integers Contains the original list past by the user; this needs to be stored since empty list in ‘ij’ are overwritten.
- read(fh, **kwargs)[source]¶
Read the two-site density matrices.
Arguments
- fhfilehandle
Open file with results. The next lines contain the measurements of the correlation matrices.
- paramdictionary (as keyword argument)
Contains the settings for the simulation.
- dynamicbool (as keyword argument)
Flag if dynamics (True) or statics (False) are read.
- ldint (as keyword argument)
Local dimension of the Hilbert space.
- class obsterms.RhoredObservable(Ops)[source]¶
Defines the measurement of a reduced density matrix of an arbitrary subsystem
Details
The arguments of the
obsterms.Observables.AddObservable()are- termtypestr
Identification as an arbitrary reduced density matrix must be ‘DensityMatrix_red’.
- termlist of integers
All sites to be included in the reduced density matrix. Python indexing starting at 0 for first site.
- maxdimint, optional
Prevents memory problems by avoiding larger reduced density matrices setting a maximal size of the total Hilbert space of the subsystem. Default to 4096 (corresponds to 12 qubits).
Example
>>> Obs = mps.Observables(Operators) >>> Obs.AddObservables('DensityMatrix_red', [0, 1, 2])
Indices must be sorted ascending. The corresponding dictionary key for accessing the reduced density matrix is the tuple (‘rho’, 0, 1, 2).
Information
The quick estimate is that the permutation of the indices to be kept followed by a partial trace has a reasonable performance. For this reason, the reduced density matrix involves truncations induced by the swapping of sites.
Developers note
The swapping scales with
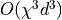 . The contraction
of the reduced density matrix can be done completely within the
ket-state (
. The contraction
of the reduced density matrix can be done completely within the
ket-state (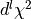 ) except the last contraction of
the ket with the bra (
) except the last contraction of
the ket with the bra (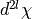 ), where
), where 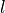 is
the dimension of the subsystem of the reduced density matrix.
is
the dimension of the subsystem of the reduced density matrix.If we do not permute sites, we have to trace out intermediate sites, which pushes the contraction of bra-ket states to the front of the algorithm. The contraction of the reduced density matrix up to site k is contracted with the ket of site (k+1) followed by the contraction of the bra of site (k+1) at costs of
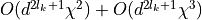 ,
where
,
where 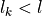 contains the number site remaining in
the reduced density matrix up to now. The final contraction
of the l-th site can use orthogonality and scales with
contains the number site remaining in
the reduced density matrix up to now. The final contraction
of the l-th site can use orthogonality and scales with
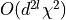 .
.Thus, we consider the swapping method as better scaling despite the introduction of an error and possible overhead for doing a significant amount of swapping operations.
- class obsterms.MutualInformationObservable[source]¶
Measurement of the mutual information matrix.
Details
The arguments of the
obsterms.Observables.AddObservable()are- termtypestr
‘MI’ to signal mutual information.
- termbool
Status of the site entropy measurement’
Truefor measuring,Falsefor not measuring.
Example
By default the mutual information is not measured. The measurement can be turned on by calling the
obsterms.Observables.AddObservable()function as follows.>>> Obs = mps.Observables(Operators) >>> Obs.AddObservable('MI', True)
Replacing
TruewithFalse, the site entropy can be turned off.Variables
The following class variables are defined.
- hasbool
Falseif site entropy should not be saved,Trueotherwise measuring the site entropy at every splitting.
- add(term, *args, **kwargs)[source]¶
Add functions as method to turn site entropy measure on and off. Last call determines if the site entropy is measured or not. For arguments see
obsterms.SiteEntropyObservable
- read(fh, **kwargs)[source]¶
Read the results from file. The filehandle must be at the corresponding position.
Arguments
- fhfilehandle
Open file with results. The next lines contain the measurements of the Schmidt decomposition.
- class obsterms.LambdaObservable[source]¶
Lambda observable taking care of the measurements of the singular values of the reduced density matrices of the bipartitions.
Details
The arguments of the
obsterms.Observables.AddObservable()are- termTypestr
string must be
Lambda.- termlist
Empty list to measure at all splittings or specifying the splittngs to be saved.
Example
In order to indicate the measurement of the singular values, you call
obsterms.Observables.AddObservable()as follows:>>> Obs = mps.Observables(Operators) >>> Obs.AddObservable('Lambda', [])
Variables
- hasbool
Falseif no singular values should be saved,Trueotherwise.- Listlist of indices
contains the list with the indices of bipartitions to be measured.
- Origlist of indices
contains the original list passed as argument. This original list is references to generate the list for each simulations. This is important in case of an empty list where the final argument depends on the system size and may vary.
- add(term, *args, **kwargs)[source]¶
Add term for measuring the singular values. Can only be called once. For arguments see class description
obsterms.LambdaObservable
- all_values(ll)[source]¶
Iterate over all possible values which can be measured.
Arguments
- llint
system size
- read(fh, **kwargs)[source]¶
Read the results from file. The filehandle must be at the corresponding position.
Arguments
- fhfilehandle
Open file with results. The next lines contain the measurements of the Schmidt decomposition.
- read_hdf5(gh5, key, **kwargs)[source]¶
Read the results from an open HDF5 file.
Arguments
- fhid of HDF5 group
This is the HDF5 group containing the observable.
- class obsterms.DistancePsiObs[source]¶
DistanceObs measures the distance to a pure state.
Details
- termlist of strings
Contains the filename containing the state to measure the distance to.
- namestr, keyword argument
The key to access the result in the result dictionary. Must be present.
- diststr, optional
Either
TraceorInfidelityto specify which quantum distance is used. Default toInfidelity.
Example
>>> Obs = mps.Observables(Operators) >>> Obs.AddObservables('dist_psi', 'somempsstate.bin', name='distpsi')
Variables
- termlist of strings
Contains the filename containing the state to measure the distance to.
- namelist of strings
Keys for access in the result dictionary.
- add(term, *args, **kwargs)[source]¶
Add a new distance measurement. The type of distance measure can be choosen.
Arguments
- termstr
Contains the filename containing the state to measure the distance to.
- diststr
The type of distance measures used. Can be
TraceorInfidelity.- namestr
String identifier to address measurement in result dictionary.
- class obsterms.StateObservable[source]¶
Write out the state as file to have access to it lateron.
Details
The arguments of :py:func:obsterms.Observables.AddObservable` for setting the state observable are
- termbool
Status of writing the state.
Truefor writing it to file,Falsefor not measuring.- formcharacter, optional
Output format of the file which can be binary ‘B’ or ‘H’ for human readable text. Default to ‘B’ (binary).
Example
By default the states are not written to file as this measurement can be memory intensive. In order to set this measurement, follow
>>> Obs = mps.Observables(Operators) >>> Obs.AddObservable('State', True)
Replacing
TruewithFalse, the measurement can be turned off again. (Currently not used in ED.)Variables
The following class variables are defined.
- hasbool
Falseif states are not written out.Trueotherwise.- file_formatstr
Defines if binary or text is written.
- add(term, *args, **kwargs)[source]¶
Add functions as method to turn state measurements on and off. Last call determines if the site entropy is measured or not. For arguments see
obsterms.StateObservable
- read(fh, **kwargs)[source]¶
This result is not stored in the result file, but a different file. Therefore, read is empty. (Still have to make it an iterator.)
- class obsterms.ETHObservable(rhoijk)[source]¶
Check the eigenstate thermalization hypothesis (ETH) for a subsystem. This measurement is calculated a posteriori within the python frontend based on the reduced density matrix. At present, it is for the first sites of the system.
Variables
The arguments of the
obsterms.Observables.AddObservable()are- termtypestr
Identification as ETH measurement must be ‘ETH’.
- terminteger
ETH is checked for the first site until site
term.- Haminstance of MPO
The Hamiltonian of the system. The rule sets are used to build a truncated Hamiltonian on the subsystem.
- kBfloat, optional
Value of Boltzmann constant. Default to 1.0
- maxdimint, optional
Prevents memory problems by avoiding larger reduced density matrices and diagonalization of the Hamiltonian of the same size. Default to 4096 (corresponds to 12 qubits).
- SymmSecNone or instance of SymmetrySector, optional
If symmetry sector is passed, ETH is checked in the corresponding sector. As the symmetries in OSMPS are global symmetries, a corresponding symmetry in a subsystem only occurs in special cases.
- sparsebool, optional
Control for using sparse matrices when building the Hamiltonian. Default to dense matrices.
Example
>>> Obs = mps.Observables(Operators) >>> Obs.AddObservables('ETH', 10)
- read(fh, **kwargs)[source]¶
Nothing written on fortran side. So read is empty. See
getfor how to obtain results.
- class obsterms.GateObservable[source]¶
Measure the overlap between state
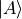 and state
and state
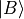 , where a series of gates
, where a series of gates 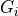 is applied to
is applied to
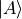 to obtain
to obtain 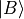 .
.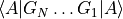
At present this feature is only supported for pure states in the EDLib and two site gates.
Variables
- Gateslist
Defines the quantum circuits.
- namelist of strings
String identifier of the meaurement.
- weightlist of floats
Weight for the each measurement.
Details
In order to add a new gate measurement, call
obsterms.Observables.AddObservable()with the following arguments:- termTypestr
string must be
Gates- termtuple / list
The first entry contains the list of gates/matrices, the second contains the list of indices.
- namestr (optional keyword argument)
Identifier for this measurement in the results.
- weightfloat (optional keyword argument)
Overall weight for measurement.
- add(term, *args, **kwargs)[source]¶
Add a new gate measurement consisting of a list of gates and the corresponding indices where it should be applied.
Arguments
- termlist/tuple of np 2d arrays; single np 2d array
d^m x d^m matrices which are the gates to be applied to the state. They should be defined on the local Hilbert spaces.
- idxlist/tuple of integers; single int
specifies the left-most site where the gate should be applied.
- namestr (optional keyword argument)
Identifier for this measurement in the results.
- weightfloat (optional keyword argument)
Overall weight for measurement.
- class obsterms.DistanceRhoObs[source]¶
DistanceObs measures the distance to another density matrix.
Variables
- termlist of strings
Contains the filename containing the state to measure the distance to.
- namestring
Label for the distance measurement. Required.
- distlist of strings
The type of distance measures used. Can be
TraceorInfidelity. Default toInfideltiy.
- add(term, *args, **kwargs)[source]¶
Add a new distance measurement. The type of distance measure can be choosen.
Arguments
- termstr
Contains the filename containing the state to measure the distance to.
- diststr
The type of distance measures used. Can be
TraceorInfidelity.- namestr
String identifier to address measurement in result dictionary.
- class obsterms.Observables(Operators)[source]¶
-
- SpecifyCorrelationRange(corr_range)[source]¶
Set the range of the correlation functions for an iMPS simulation
Arguments
- corr_rangeInteger
distance for the correlation measurement in a iMPS simulation.
- read_imps(fh, param)[source]¶
Read the results of an iMPS simulation for one convergence parameter set. The possible degenerate states are accessible as a nested dictionary, key is
('DegMeas', ii)for the ii-th degenerate state. The values ofii = 0are present in the usual result dictionary.Arguments
- fhfilehandle
open filehandle at correct position to read iMPS results.
- paramdictionary
Simulation setup as dictionary to retrieve settings.
- write(filestub, Parameters, IntOperators, IntHparam, PostProcess=False)[source]¶
Write the Observables object to a Fortran-readable file.
Arguments
- fileStubstr
basis part of the filename for this simulation extended with
Obs.dat.- Parametersdictionary
dictionary representing the simulation to be written.
- IntOperatorsdictionary
mapping the string identifiers for the operators to integer identifiers used within fortran. Needed to write MPO measurements.
- IntHparamdictionary
mapping the string identifiers for Hamiltonian parameters to integer identifiers used within fortran. Needed to write MPO measurements.
- PostProcessbool, optional
The fortran files are only written if
False. Otherwise only the necessary data is updated, e.g. the list containing the settings for the reduced density matrices. Default toFalse
- class obsterms.Result(param, case, state=0, conv_ii=1, FiniteT=False)[source]¶
Arguments
- paramdictionary
parameter dictionary setting up a single simulation
- casestr
Specifies the measurement, either ‘MPS’ (ground state), ‘eMPS’ (excited state), ‘dynamics’ (time evolution).
- stateinteger, optional
select between the states, especially for excited states. For dynamics, store the time step in there. default to 0
- conv_iiinteger, optional
selects i-th element in the set of convergence parameters counting from 1 on. It is taken into account by an offset that python indices start at 0. For dynamics, store the number of the quench in there. default to 1
- dynamiclogical, optional
Layout of the output differs from static to dynamic. Specify dynamic=True for reading dynamic simulations. default to False
- FiniteTlogical, optional
To treat different formatting on convergence parameters in regard to Finite T simulations. Default to False.
- add_qt(dic, dicjj)[source]¶
Combine results for quantum trajectories.
Arguments
- dicdictionary
Target dictionary with the overall results.
- dicjjdictionary
Dictionary with the results of one trajectory.
- obsterms.ReadStaticObservables(Parameters)[source]¶
Read the observables written out by Fortran program.
Arguments
- Parameterslist
list of dictionaries where the dictionaries contain the simulation settings.
Details
For infinite simulations see
ReadInfiniteStaticObservables(). The finite results consists of a list of dictionaries. Each dictionary corresponds to one simulation. Simulation which could not be read (missing/incomplete files) can either throw an exception or deliverNoneas entry in the list.The default static finite system measurements are listed in the following tables. In addition the measurments defined in
obsterms.Observablesare available:Tag
Meaning
state
Eigenstate index increasing in energy with 0 the ground state.
convergence_parameter
Integer index indicating convergence criteria used.
energy
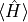
variance
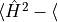
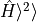
converged
T (True) or F (False) according to whether the variance satisfies the desired tolerance.
bond_dimension
The maximal bond dimension of the MPS.
The default static infinite system measurements can be found in the corresponding class description
obsterms.ResultsiMPS. An instance of this class is returned.Errors in the process of reading can be catched upon the first access to the any measurement.
ReadStaticObservablesjust generates an array of the result class, which will process the requests to access results. An error handling could look as follows>>> Outputs = mps.ReadStaticObservables(params) >>> energies = [] >>> >>> for elem in Outputs: >>> try: >>> print(Outputs['energy']) >>> energies.append(Outputs['energy']) >>> except: >>> print('Error processing result.') >>> energies.append(np.nan)
The list energies has the expected length after reading the results and non-existent values are set with nan (not a number).
- obsterms.ReadDynamicObservables(Parameters)[source]¶
Read the observables written out by Fortran program.
Arguments
- Parameterslist
list of dictionaries where every dictionary contains the simulation setting.
Details
The default dynamic measurements are listed in the following tables. In addition the measurments defined in
obsterms.Observablesare available:Tag
Meaning
time
current time
energy
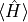
Loschmidt_Echo
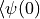
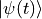
bond_dimension
The maximal bond dimension of the MPS.
Errors in the process of reading can be catched upon the first access to the any measurement.
ReadDynamicObservablesjust generates an array of the result class, which will process the requests to access results. An error handling could look as follows>>> Outputs = mps.ReadDynamicObservables(params) >>> energies = [] >>> >>> for elem in Outputs: >>> try: >>> print(Outputs['energy']) >>> energies.append(Outputs['energy']) >>> except: >>> print('Error processing result.') >>> energies.append(np.nan)
The list energies has the expected length after reading the results and non-existent values are set with nan (not a number).
- obsterms.ReadCircuitsObservables(elem)[source]¶
Read the observables from a quantum circuit simulation.
- obsterms.GetObservables(Outputs, tags, values)[source]¶
Find the set of all Outputs which have the values specified in tags.
Arguments
- Outputslist of results
This is a list containing all result dictionaries.
- tagsvariables or list of variables
These are the keys, i.e., the parameters you use for selection.
- valuesvariable, list of variables
Values for the keys in the same order and of same length if list.
- obsterms.get_runtime(param, qtid=None)[source]¶
Read runtime from log-file.
Arguments
- paramdict
dictionary with simulation setup.
- class obsterms.OSMPS_H5Method_Error(ret_val)[source]¶
Excpetion raised if an OSMPS method is not enabled for H5File at all, but was trying to read with HDF5.
Arguments __init__
- ret_valstr
Description of the case.
- class obsterms.OSMPS_H5File_Error(ret_val)[source]¶
Excpetion raised if an H5File does not exist.
Arguments __init__
- ret_valstr
The filename which does not exist
- class obsterms.OSMPS_H5Group_Error(ret_val)[source]¶
Exception raised if an H5 group does not exist.
Arguments __init__
- ret_valstr
Name of the group.