Python Module exactdiag¶
exactdiag.py Module defines a pure state and a density matrix based on a general abstract quantum system. It defines time evolution methods for those systems.
- exactdiag.set_spectrum(val)[source]¶
Set the value for the calculation of the spectrum.
Arguments
- valboolean
For calculating the spectrum need
True, otherwiseFalse.
- exactdiag.runED(params, Operators, Hmpos, sparse=True, maxdim=None)[source]¶
Run all simulation in the list built of parameter dictionaries.
Arguments
- paramslist
list of dictionaries where each dictionaries represents a MPS simulation.
- Operatorsinstance of
Ops.OperatorList(or dictionary) Operators defined used in the simulation.
- Hmposinstance of
MPO.MPOor list ofMPO.MPO Hamiltonian represented in the MPO class.
- sparsebool, optional
Flag on the use of sparse matrices. Default to True (use sparse matrices).
- maxdimNone or int, optional
Maximal dimension for the complete Hilbert space in order to prevent to large systems for ED. Default to None (no limitation).
- exactdiag.runED_threading(params, Operators, Hmpos, threads, sparse=True, maxdim=None, tsleep=30.0)[source]¶
Allows to run ED simulations with threading on a single node.
Arguments
- paramslist
list of dictionaries where each dictionaries represents a MPS simulation.
- Operatorsinstance of
Ops.OperatorList(or dictionary) Operators defined used in the simulation.
- Hmposinstance of
MPO.MPOor list ofMPO.MPO Hamiltonian represented in the MPO class.
- sparsebool, optional
Flag on the use of sparse matrices. Default to True (use sparse matrices).
- maxdimNone or int, optional
Maximal dimension for the complete Hilbert space in order to prevent to large systems for ED. Default to None (no limitation).
- tsleepfloat, optional
Time interval to check for finished jobs.
Details
The multiprocessing module should be taking care to obtain thread-safe processes, but complications might arise from there.
- exactdiag.ed_simulation(Hmpo, Operators, param, sparse, maxdim, qtid)[source]¶
Carry out a single static and dynamics simulations for one single set of parameters.
Arguments
- Hmpoinstance of
MPO.MPO Hamiltonian represented in the MPO class.
- Operatorsinstance of
Ops.OperatorList(or dictionary) Operators defined used in the simulation.
- paramdictionary
dictionary representing a MPS simulation.
- sparsebool
Flag on the use of sparse matrices.
- maxdimint
Maximal dimension for the complete Hilbert space in order to prevent to large systems for ED.
- qtidinteger
index for multiple realizations of the same simulations, e.g. with quantum trajectories.
- Hmpoinstance of
- exactdiag.dynamic_simulation(Psi, Psi0, Hmpo, Operators, param, sparse, maxdim, qtid)[source]¶
Run the dynamics for a single parameter set.
Arguments
- PsiPure state or density matrix.
Quantum state for time evolution. It can either be represented as instance of
exactdiag.PureStateor as instance ofexactdiag.DensityMatrix- Psi0instance of
exactdiag.PureState Quantum state to calculate overlap with the state at every measurement. Meant to be used with the state at time t=0 for the Loschmidt echo.
- Hmpoinstance of
MPO.MPO Hamiltonian represented in the MPO class.
- Operatorsinstance of
Ops.OperatorList(or dictionary) Operators defined used in the simulation.
- paramdictionary
dictionary representing a MPS simulation.
- sparsebool
Flag on the use of sparse matrices.
- maxdimint
Maximal dimension for the complete Hilbert space in order to prevent to large systems for ED.
- qtidinteger
If > 0, Psi as an instance of
exactdiag.PureStateis evolved with quantum trajectories (open system). If 0, instance ofexactdiag.PureStateis evolved under the Schrodinger equation. If Psi is an instance ofexactdiag.DensityMatrix, it is evolved in the Liouville space with the Lindblad master equation and the flag qtid has no effect.
- exactdiag.dynamic_circuits(Psi, Psi0, Operators, param, QCin=None)[source]¶
Evolve the quantum system with circuits.
Arguments
- PsiPure state or density matrix.
Quantum state for time evolution. It can either be represented as instance of
exactdiag.PureStateor as instance ofexactdiag.DensityMatrix- Psi0instance of
exactdiag.PureState Quantum state to calculate overlap with the state at every measurement. Meant to be used with the state at time t=0 for the Loschmidt echo.
- Operatorsinstance of
Ops.OperatorList(or dictionary) Operators defined used in the simulation.
- paramdictionary
dictionary representing a MPS simulation.
- callerinstance of QuantumCircuits or None, optional
If QCin is not
None, measurements are skipped and the final state is returned. Default toNone.
- exactdiag.expm_symm_tridiag(diag, sdiag, sc)[source]¶
Calculate exponential of symmetric tridiagonal matrix.
Arguments
- diag1d numpy array
diagonal entries (dimension of the matrix)
- sdiag1d numpy array
supdiagonal entries
- scfloat, complex
scaling inside of the exp(.)
- class exactdiag.SymmetrySector(Operators, param, sectorbase=None, empty=False)[source]¶
Taking care of the symmtry. Generates all basis states in the Hilbert space and returns the indices of the states which are conform with the quantum numbers defined for the simulations. The result returned is a list of integers, unless there are no symmetries defined (None).
Arguments
- Operatorsinstance of
Ops.OperatorList Contains all operators of the simulation, especially the diagonal generators of the symmetry.
- paramdictionary
Contains the simulation setup.
- sectorbase2d numpy array, optional
If basis states are calculated manually, a 2d numpy array with a basis state in each row can be passed to initialize the symmetry sector. Number of rows is the number of basis states, number of columns corresponds to the number of sites. For
Operatorsandparamare not referenced andNonecan be passed to them. Default to None.- emptybool, optional
For internal use, no variables are set. Allows for setting them manually. Default to False
Variables
- lenint
number of basis states in the symmetry sector.
- sectorbase2d numpy array, str
local basis for each basis state of the symmetry sector. If str, filename of the 2d numpy array to be loaded.
- sectordicdict
For a tuple with the local basis state the corresponding index is returned. Does not handle key errors.
- emptybool, optional
for internal use only. Default to False
- __call__(ii)[source]¶
Return the indices of the local basis for the ii-th state.
Arguments
- iiint
Consider to ii-th state in the symmetry-sector based basis.
- __getitem__(key)[source]¶
Getitem is defined to return the index of a basis state.
Arguments
- keylist, array (1d)
contains the local basis for each site
- get_subspace(idxs)[source]¶
Return the symmetry adapted basis of a subspace.
Arguments
- idxslist of integers
these are the sites to be considered for the new space.
- Operatorsinstance of
- exactdiag.walls_particles(ll, fill)[source]¶
The distribution of fill particles on ll sites can be described via fill particles and (ll - 1) walls. This iterator yields all possible combinations where 0s represent a wall and 1s represent the a particle.
Arguments
- llint
number of sites in the system.
- fillint
number of particles distributed on the system.
- exactdiag.SingleU1Symmetry(Operators, param)[source]¶
Create symmetry sector for Bose-Hubbard type models with minimum number of particles zero up to some maximum filling.
Arguments
- Operatorsinstance of
Ops.OperatorList contains all operators including the generator of the symmetry. It is checked that the symmetry generator is the number operator.
- paramdictionary
Contains the simulation setup. It is checked that only one U(1) symmetry is present.
- Operatorsinstance of
- exactdiag.statics_simulation(Hmpo, Operators, param, SymmSec, sparse, maxdim)[source]¶
Complete a static simulation including finding the ground and excited states and measuring those. Returns the ground state if needed for dynamics.
Arguments
- Hmpoinstance of
MPO.MPO Hamiltonian written in terms of MPO rules.
- Operatorsinstance of the operators class
Ops.OperatorList containing the operators to generate Hamiltonian.
- paramdictionary with simulation parameters
settings of the whole simulation.
- SymmSeclist or None
list of indices containing the states in the given basis which are in the symmetry sector considered.
- sparsebool
flag if sparse matrix should be used for Hamiltonian
- maxdimint
limits the dimension of the complete Hilbert space.
- Hmpoinstance of
- exactdiag.statics_state_sparse(Hmpo, param, ii, SymmSec, maxdim)[source]¶
Find a specific state of the Hamiltonian. Returns state and energy (in this order). Function for dense Hamiltonians.
Arguments
- Hmpoinstance of
MPO.MPO Hamiltonian written in terms of MPO rules.
- paramdictionary with simulation parameters
settings of the whole simulation.
- iiint, optional
index of the state to be returned. If 0, ground state is returned, if 1 first excited state and so on. No specific handling for degenerate states. Default to 0 (ground state)
- SymmSeclist or None
list of indices containing the states in the given basis which are in the symmetry sector considered.
- maxdimint, optional
limits the dimension of the complete Hilbert space.
- Hmpoinstance of
- exactdiag.statics_state_dense(Hmpo, param, ii, SymmSec, maxdim)[source]¶
Find a specific state of the Hamiltonian. Returns state and energy (in this order). Function for dense Hamiltonians.
Arguments
- Hmpoinstance of
MPO.MPO Hamiltonian written in terms of MPO rules.
- paramdictionary with simulation parameters
settings of the whole simulation.
- iiint, optional
index of the state to be returned. If 0, ground state is returned, if 1 first excited state and so on. No specific handling for degenerate states. Default to 0 (ground state)
- SymmSeclist or None
list of indices containing the states in the given basis which are in the symmetry sector considered.
- maxdimint, optional
limits the dimension of the complete Hilbert space.
- Hmpoinstance of
- class exactdiag._QuantumSystem[source]¶
Abstract type for a quantum system, either pure state or density matrix. Implements general methods equal for both.
- apply_op(Op, idx)[source]¶
Apply an operator or gate for any number of sites to the quantum systems. Operatorcan either be for pure states or for density matrices, but is defined in the Hilbert space for both of them
Arguments
- Opnp 2d array
containing the operator to be applied, e.g. the propagator of a Hamiltonian or Liouvillian.
- idxlist of integers
Apply term to the sites specified here.
- apply_op2(Op, ii, ll_1_term=False, perm=False)[source]¶
Apply an operator for two neighboring sites to the quantum system. Operator can either be for pure states or density matrix (Liouville space).
Arguments
- Opnp 2d array
containing the operator to be applied, e.g. the propagator of a Hamiltonian or Liouvillian.
- iiint
Apply term to the sites ii and (ii + 1).
- ll_1_termbool, optional
Boolean to indicate the term ii=L for periodic boundary conditions. Default to
False.
- dot(*args)[source]¶
General dot product split into different cases, which are detected by the data type.
- getset_rho(key)[source]¶
Return the reduced density matrix from the complete wave function.
Arguments
- keytuple
containing one or two integers specifying the site indices.
- meas_observables(Operators, param, fh, obstype)[source]¶
Measure the observables.
Arguments
- Operatorsinstance of
Ops.OperatorList All operators defined for the simulation.
- paramdict
containing simulation setup, necessary for MPO measurement.
- fhfilehandle
Open file. Results are written to this file handle.
- obstypestring
Key to get Observables class in param.
- Operatorsinstance of
- propagate(dt, Hmpo, Operators, param, sparse, maxdim, quenchtime, qt)[source]¶
Time evolution of a quantum system for one time step with either of the following three methods: matrix exponential, Krylov approximation to the matrix exponential, or Trotter decomposition. Method is determined via the convergence parameters.
Arguments
- dtfloat
time step (discretization of time).
- Hmpoinstance of
MPO.MPO Defines the Hamiltonnian of the system.
- Operatorsinstance of
Ops.OperatorList Contains the operators used to build the Hamiltonian.
- paramdictionary
Setup for the simulation.
- sparsebool
Flag for the use of sparse methods (only referenced for matrix exponential method and (possibly) Krylov).
- maxdimint
maximal dimension of the Hilbert space. Only referenced in …
- quenchtimetuple of int and float
Storing the number of the quench and the actual time
- qtboolean
If
True, this as an instance ofexactdiag.PureStateis evolved with quantum trajectories (open system). IfFalse, instance ofexactdiag.PureStateis evolved under the Schrodinger equation. If Psi is an instance of aexactdiag.DensityMatrix, it is evolved in the Liouville space with the Lindblad master equation and the flag qt has no effect.
- propagate_trotter(dt, Hmpo, Operators, param, quenchtime, qt)[source]¶
Propagate the quantum system with a Trotter decomposition. This function calls depending on the order of the decomposition the
exactdiag.PureState.trotter_layer()orexactdiag.DensityMatrix.trotter_layer()for the odd/even part.Arguments
- dtfloat
size of the time step/discretization.
- Hmpoinstance of
MPO.MPO Hamiltonian as rule set for the Trotter decomposition.
- Operatorsinstance of
Ops.OperatorList(or dictionary) Operators defined used in the simulation.
- paramdictionary
Contains the simulation setup.
- quenchtimetuple of int and float
Storing the number of the quench and the actual time
- qtbool
If
Truequantum states are evolved under quantum trajectories, else (False) quantum states are evolved under the Schrodinger equation. For DensityMatrix this should be alwaysFalse.
Details
The default order for the Trotter decomposition is the 2nd order. The default is overwritten with the order in
convergence.TEBDConvParamif this class is given. Possible values are therefore 2 and 4.
- reduced(keep, enforce_dense=False)[source]¶
Build the reduced density matrix of the system for a specified set of sites.
Arguments
- keeplist of int, optional
Site indices to be kept in reduced density matrix. Must be between 0 < ll - 1 and unique. Consistency with trce is not checked. Default to None (indices are generated from trce)
- enforce_densebool, optional
Return reduced density matrix on complete Hilbert space without using symmetries (True). Otherwise symmetries might be used if considered convenient. Default to False.
- save_propagator_spectrum(Mat, param, quenchtime, tol=1e-13)[source]¶
Save the eigenvalues of the spectrum of the Hamiltonian or Liouville operator.
Arguments
- Mat2d matrix
Contains the Hamiltonian or Liouville operator with the eigenvalues to be determined.
- paramdictionary
Contains the simulation setup for one simulation.
- quenchtimetuple of integer and float
Contains the number of the corresponding quench and the current time of the simulation.
- tolfloat, optional
Tolerance for the check of hermiticity. Default to 1e-13.
Details
- traceout(trce, keep=None, enforce_dense=False)[source]¶
Carry out the partial trace on the system returning the density matrix for the remaining system.
Arguments
- trcelist of int
Site indices to trace out. Must be between 0 < ll - 1 and unique.
- keeplist of int, optional
Site indices to be kept in reduced density matrix. Must be between 0 < ll - 1 and unique. Consistency with trce is not checked. Default to None (indices are generated from trce)
- enforce_densebool, optional
Return reduced density matrix on complete Hilbert space without using symmetries (True). Otherwise symmetries might be used if considered convenient. Default to False.
- trotter_Ham_2site(Hmpo, Operators, param, quenchtime, even)[source]¶
Build the Hamiltonian for 2 neighboring site containing only local and bond rules.
Arguments
- Hmpoinstance of
MPO.MPO Defines the Hamiltonnian of the system.
- Operatorsinstance of
Ops.OperatorList Contains the operators used to build the Hamiltonian.
- paramdictionary
Setup for the simulation.
- quenchtimetuple of int and float
Storing the number of the quench and the actual time
- evenbool
Boolean to switch between even (
True) and odd (False) layers in the Trotter decomposition.
- Hmpoinstance of
- class exactdiag.PureState(Psi, ld, ll, SymmSec)[source]¶
Pure state vector for the evolution of closed systems.
Arguments
Psi : 1d numpy array
- ldint
local dimension in the Hilbert space
- llint
number of sites in the system
- SymmSecNone or instance of
exactdiag.SymmetrySector contains the setup of the symmetry if present in the system.
Variables
- ddint
dimension of Psi
- rhosdict
Storing reduced density matrices for measurements.
- lastpropNone, 2d numpy array, or dictionary.
Last propagator used for time independent quenches either as numpy 2d array for the matrix exponential or as dictionary for the Trotter decomposition.
- bidxdictionary
Used to store the indices of the subblocks with the same symmetry numbers.
- _apply_op2_nosymm(Op, ii, ll_1_term, perm)[source]¶
Apply a two-site operator to the state vector on a specified site.
Arguments
- Opnp 2d array
The operator acting on two sites represented as d^2 x d^2 matrix.
- iiint
Apply term to the sites ii and (ii + 1).
- ll_1_termbool, optional
Boolean to indicate the term ii=L for periodic boundary conditions.
- _apply_op2_symm(Op, ii, ll_1_term, *args)[source]¶
Apply a two-site operator to the state vector on a specified site.
Arguments
- Opnp 2d array
The operator acting on two sites represented as d^2 x d^2 matrix.
- iiint
Apply term to the sites ii and (ii + 1).
- ll_1_termbool, optional
Boolean to indicate the term ii=L for periodic boundary conditions.
- _apply_op_nosymm(Op, idx)[source]¶
Apply an operator/gate on an arbitrary number of sites.
Arguments
- Opnp 2d array
The operator acting on the site represented as matrix.
- idxlist of integers
The sites to be contracted with the operator.
- _apply_op_symm(Op, idx)[source]¶
Apply an operator/gate on an arbitrary number of sites to a symmetry conserving state.
Arguments
- Opnp 2d array
The operator acting on the site represented as matrix.
- idxlist of integers
The sites to be contracted with the operator.
- _bond_entropy_to_left()[source]¶
Start from ii = ll / 2 and calculate bond entropies going to the left.
- _bond_entropy_to_right()[source]¶
Start from ii = ll / 2 + 1 and calculate bond entropies going to the left.
- _dot_effhmpo_vec_nosymm(Hmpo, Operators, param, quenchtime, maxdim)[source]¶
Multiply the effective Hamiltonian represented as MPO / MPO rule set with a vector. For arguments see
exactdiag.PureState.dot_effhmpo_vec(). Intended for internal use.
- _dot_effhmpo_vec_symm(Hmpo, Operators, param, quenchtime, maxdim)[source]¶
Multiply the effective Hamiltonian represented as MPO / MPO rule set with a vector. The vector is defined on the symmetry sector specified. For arguments see
exactdiag.PureState.dot_effhmpo_vec(). Intended for internal use.
- _dot_hmpo_vec_nosymm(Hmpo, Operators, param, quenchtime, maxdim)[source]¶
Multiply a Hamiltonian represented as MPO / MPO rule set with a vector. For arguments see
exactdiag.PureState.dot_hmpo_vec(). Intended for internal use.
- _dot_hmpo_vec_symm(Hmpo, Operators, param, quenchtime, maxdim)[source]¶
Multiply a Hamiltonian represented as MPO / MPO rule set with a vector. The vector is defined on the symmetry sector specified. For arguments see
exactdiag.PureState.dot_hmpo_vec(). Intended for internal use.
- _get_bond_entropy_nosymm(ii)[source]¶
Calculate the bond entropy for a state of splitting (0, …, ii), (ii + 1, …, L - 1) in a system where no symmetries are used. For description of other arguments look into
get_bond_entropies.Arguments
- iiint
Specifies the index of the splitting, where ii is the last site included in the left bipartition.
Notes
Could use scipy.linalg.blas.zherk or scipy.linalg.blas.dsyrk
- _get_bond_entropy_symm(ii)[source]¶
Calculate the bond entropy for a state of splitting (0, …, ii), (ii + 1, …, L - 1) in a system with symmetries. For description of arguments look into
get_bond_entropies.Arguments
- iiint
Specifies the index of the splitting, where ii is the last site included in the left bipartition.
- _matrix_exp_trotter(Ham)[source]¶
Calculate the matrix exponential considering block diagonal structure if symmetries are present.
Arguments
- Hamnp 2d array
matrix representing the Hamiltonian including the time step. To be exponentiated.
Details
Functions accesses
self.bidx, a dictionary of tuples orNone. Each set of tuples contains the indices for the same symmetry sector and the corresponding site indices.
- _quantum_jump_nosymm(Hmpo, param, quenchtime)[source]¶
Apply a quantum jump for the quantum trajectory evolution in a system without any symmetry present. For arguments look into documentation of
exactdiag.PureState.quantum_jump(). Intended for internal use.
- _quantum_jump_symm(Hmpo, param, quenchtime)[source]¶
Apply a quantum jump for the quantum trajectory evolution in a system with one or multiple symmetries present. For arguments look into documentation of
exactdiag.PureState.quantum_jump(). Intended for internal use.
- _traceout_nosymm(trce, keep)[source]¶
Trace over a set of sites specified via their indices for states without a symmetry.
Arguments
- trcelist of ints
Trace out over the indices contain in this list.
- keeplist of ints
Keep the sites whose indices are listed here. Together trce and keep should list all indices exactly one time.
- _traceout_symm(trce, keep, SubSec=None)[source]¶
Trace over a set of sites specified via their indices for states with symmetry returning a density matrix considering the symmetry in the larger Hilbert space.
Arguments
- trcelist of ints
Trace out over the indices contain in this list.
- keeplist of ints
Keep the sites whose indices are listed here. Together trce and keep should list all indices exactly one time.
- SubSecinstance of
exactdiag.SymmetrySector, optional If symmetry sector is already calculated, it can be passed with this argument. Otherwise pass
None. Default toNone.
- _traceout_symm2nosymm(trce, keep, *args, **kwargs)[source]¶
Trace over a set of sites specified via their indices for states with symmetry returning a density matrix not considering the symmetry.
Arguments
- trcelist of ints
Trace out over the indices contain in this list.
- keeplist of ints
Keep the sites whose indices are listed here. Together trce and keep should list all indices exactly one time.
*args,**kwargskeeping syntax with corresponding method for density matrices.
- _trotter_finalize(qt, Hmpo, param, quenchtime)[source]¶
At the end of the Trotter step, the rank-L tensor has to be converted back into the state vector if no symmetry is present. For quantum trajectories it is checked if a jump has to be applied.
Arguments
- qtbool
Trueif evolution with quantum trajectories, otherwiseFalse.- Hmpoinstance of
MPO.MPO Hamiltonian represented in the MPO class.
- paramdictionary
dictionary representing a MPS simulation.
- quenchtimetuple of int and float
Storing the number of the quench and the actual time.
- _trotter_initialize(qt)[source]¶
The initialization transfers the vector into a rank-L tensor if no symmetries are present. Furthermore the norm and random number for the quantum trajectories are created before the first time step.
Arguments
- qtbool
Trueif evolution with quantum trajectories, otherwiseFalse.
- distance(State, dist)[source]¶
Measure the distance between the pure state and another quantum state.
Arguments
- State1d or 2d numpy array
Represents another pure state (1d) or a density matrix (2d).
- distancestr
Define the type of distance measure. Either
TraceorInfidelity.
- dot_effhmpo_vec(Hmpo, Operators, param, quenchtime, maxdim)[source]¶
Multiply the effective Hamiltonian represented as MPO / MPO rule set with a vector.
Arguments
- Hmpoinstance of
MPO.MPO defines the effective Hamiltonian through a rule set. The open system part contributes -0.5j Ldagger L (where the imaginary unit is cancelled out when mulitplying with (-1j) in the time evolution.
- Operatorsinstance or
Ops.OperatorList containing all operators needed defined on a local Hilbert space.
- paramdictionary
setup for the simulation
- quenchtimetuple or None
If tuple containing index of quench and present time to obtain coupling.
- maxdimint
maximal dimension of the Hilbert space
- Hmpoinstance of
- dot_hmpo_vec(Hmpo, Operators, param, quenchtime, maxdim)[source]¶
Multiply a Hamiltonian represented as MPO / MPO rule set with a vector.
Arguments
- Hmpoinstance of
MPO.MPO defines the Hamiltonian through a rule set.
- Operatorsinstance or
Ops.OperatorList containing all operators needed defined on a local Hilbert space.
- paramdictionary
setup for the simulation
- quenchtimetuple or None
If tuple containing index of quench and present time to obtain coupling.
- maxdimint
maximal dimension of the Hilbert space
- Hmpoinstance of
- dot_vec(Vec)[source]¶
Return overlap of wavefunction with other state vector
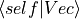 .
.Arguments
- Vecnp 1d array or instance of
exactdiag.PureState Other vector in the overlap with the PureState.
- Vecnp 1d array or instance of
- entropy()[source]¶
Calculate the von-Neumann entropy of the system (always zero for the pure state).
- fidelity(Phi)[source]¶
Return the fidelity between this state Psi and Phi defined as
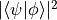 .
.Arguments
- Phinp 1d array or instance of
exactdiag.PureState Other vector in the overlap with the PureState.
- Phinp 1d array or instance of
- get_bond_entropies()[source]¶
Calculate the bond entropies for every possible splitting along the chain. Iterator returns ll + 1 numbers.
- meas_mpo(Hmpo, Operators, param, quenchtime, maxdim)[source]¶
Measure an MPO defined as rule set, e.g. the Hamiltonian.
Arguments
- Hmpoinstance of
MPO.MPO defines the Hamiltonian through a rule set.
- Operatorsinstance or
Ops.OperatorList containing all operators needed defined on a local Hilbert space.
- paramdictionary
setup for the simulation
- quenchtimetuple or None
If tuple containing index of quench and present time to obtain coupling.
- maxdimint
maximal dimension of the Hilbert space
- Hmpoinstance of
- propagate_expm(dt, Hmpo, param, sparse, maxdim, quenchtime, qt)[source]¶
Propagate a quantum state where the propagator is built from a sparse or dense Hamiltonian.
Arguments
- dtfloat
size of the time step
- Hmpoinstance of
MPO.MPO defines the Hamiltonian through a rule set.
- paramdictionary
setup for the simulation
- sparsebool
Flag on the use of sparse matrices. If
True, sparse matrices will be used.- maxdimint
maximal dimension of the Hilbert space
- quenchtimetuple
Tuple containing index of quench and present time to obtain coupling.
- qtbool
flag for quantum trajectories.
- propagate_expm_qt(dt, Hmpo, param, sparse, maxdim, quenchtime)[source]¶
Propagate a quantum state where the propagator is built from the effective Hamiltonian.
Arguments
- dtfloat
size of the time step
- Hmpoinstance of
MPO.MPO defines the Hamiltonian through a rule set.
- paramdictionary
setup for the simulation
- sparsebool
Flag on the use of sparse matrices. If
True, sparse matrices will be used.- maxdimint
maximal dimension of the Hilbert space
- quenchtimetuple or None
If tuple containing index of quench and present time to obtain coupling.
- propagate_gates(func, args=(), return_copy=False)[source]¶
Propagate a quantum state with a set of given gates. At present only 2-site gates are supported.
Arguments
- funciterator (
__iter__) return gates and left most site index where gate should be applied.
- argstuple, optional
arguments for call to the iterator. Default to ()
- return_copyboolean, optional
If
False, then the state of the class is modified/propagated. The state remains unchanged and a copy of the state after applying the sequence of gates is returned forTrue. Default toFalse.
- funciterator (
- propagate_krylov(dt, Hmpo, Operators, param, sparse, maxdim, quenchtime, qt)[source]¶
Propagate a quantum state taking the exponential in the Krylov subspace.
Arguments
- dtfloat
time step
- HmpoMPO
Rule set describing the Hamiltonian.
- Operatorsinstance of
Ops.OperatorList contains all the operators to setup the Hamiltonian
- paramdict
dictionary with simulation setup.
- sparsebool
Flag if sparse matrices should be used. Flag only used for small matrices when complete Hamiltonian is built.
- maxdimint
maximal dimension of the Hilbert space.
- quenchtimetuple or None
contains number of quench and present time.
- qtbool
flag for quantum trajectories.
Details
The following modes are available for the Krylov time evolution, which is set in the convergence criteria for the quenches, i.e.,
convergence.KrylovConvParamwith the variableedlibmode:0 : Built Hamiltonian as matrix and use built-in scipy method.
1 : Built Hamiltonian as matrix and use EDLib Krylov.
2 : Use dot product of MPO and state vector and EDLib Krylov.
3 : As 2, but save Krylov vectors to hard disk to reduce RAM memory usage.
-1 : default. For :math`dim < 2^{10}` take edlibmode=0, then for
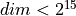 use edlibmode=2, otherwise edlibmode=3
use edlibmode=2, otherwise edlibmode=3
Since the Hamiltonian is hermitian, Krylov uses the Lanczos implementation of the algorithm.
- propagate_krylov_qt(dt, Hmpo, Operators, param, sparse, maxdim, quenchtime)[source]¶
Propagate a quantum state taking the exponential in the Krylov subspace. Although this is on pure states, we need the Krylov-Arnoldi version to account for non-hermiticity of the expression.
Arguments
- dtfloat
time step
- HmpoMPO
Rule set describing the Hamiltonian.
- Operatorsinstance of
Ops.OperatorList contains all the operators to setup the Hamiltonian
- paramdict
dictionary with simulation setup.
- sparsebool
Flag if sparse matrices should be used. Flag only used for small matrices when complete Hamiltonian is built.
- maxdimint
maximal dimension of the Hilbert space.
- quenchtimetuple or None
contains number of quench and present time.
Details
The same modes for the Krylov algorithm are available as described in
exactdiag.PureState.propagate_krylov(). Since the effective Hamiltonian is not hermitian, the Arnoldi version of the Krylov algorithm is used.
- quantum_jump(Hmpo, param, quenchtime)[source]¶
Apply a Lindblad operator for the quantum trajectory evolution. This function works for any time evolution method with an (effective) Hamiltonian (which are matrix exponential, Trotter, Krylov).
Arguments
- Hmpoinstance of
MPO.MPO contains the MPO and Lindblad operators.
- paramdict
contains the simulation setup.
- quenchtimetuple of (int, float)
information about the number of the quench and the present time to gain the coupling constants for the Lindblad operators.
- Hmpoinstance of
- trotter_layer(dt, Hmpo, Operators, param, quenchtime, even, qt)[source]¶
Apply an even or odd layer of the Trotter decomposition to the state vector.
Arguments
- dtfloat
time step/discretization of the simulation.
- Hmpoinstance of
MPO.MPO Hamiltonian represented in the MPO class.
- Operatorsinstance of
Ops.OperatorList Contains the operators used to build the Hamiltonian.
- paramdictionary
dictionary representing a MPS simulation.
- quenchtimetuple of int and float
Storing the number of the quench and the actual time.
- evenbool
Boolean to switch between even (
True) and odd (False) layers in the Trotter decomposition.- qtbool
Flag if using Schrodinger equation (
False) or quantum trajectories (True).
- trotter_layer_perm(even, ll, pbc)[source]¶
Return permutation to be applied after a complete layer of two-site Trotter terms.
Arguments
- evenbool
Indicates if the even or odd layer of the Trotter decomposition was applied.
- llint
System size.
- pbcbool
Indicates if the Hamiltonian has periodic boundary conditions or not.
- trotter_layer_qt(dt, Hmpo, Operators, param, quenchtime, even)[source]¶
Trotter layer under the effective Hamiltonian of the Lindblad master equation representing the hermitian part for an even or odd layer.
Arguments
- dtfloat
time step/discretization of the simulation.
- Hmpoinstance of
MPO.MPO Hamiltonian represented in the MPO class.
- Operatorsinstance of
Ops.OperatorList Contains the operators used to build the Hamiltonian.
- paramdictionary
dictionary representing a MPS simulation.
- quenchtimetuple of int and float
Storing the number of the quench and the actual time.
- evenbool
Boolean to switch between even (
True) and odd (False) layers in the Trotter decomposition.
- class exactdiag.DensityMatrix(Arr, ld, ll, SymmSec)[source]¶
Density matrix for evolution of open systems.
Arguments
- Arr1d or 2d np array
If 1d array, a state vector Psi is assumend and the density matrix is built as
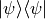 . If 2d array, array
is assumed to be the density matrix.
. If 2d array, array
is assumed to be the density matrix.- ldint
local dimension of the Hilbert space for each site.
- llint
number of sites in the system.
- SymmSec: None or instance of
exactdiag.SymmetrySector contains the setup for symmetries. If no symmetry is present, pass
None.
Variables
- Dint
Dimension of the Hilbert space
- rho2d numpy array
Density matrix
- rhosdict
Storing reduced density matrices
- lastpropNone, 2d numpy array, or dictionary.
Last propagator used for time independent quenches. If 2d numpy array, the propagator is stored as matrix. If dictionary, the local propagators for the Trotter decomposition are stored.
- bidxdictionary
Used to store the indices of the subblocks with the same symmetry numbers.
- _apply_op2_nosymm(Op, ii, ll_1_term, *args)[source]¶
Apply a two-site operator to the density matrix on a specified site. This is the version for density matrices without a symmetry adapted basis.
Arguments
- Opnp 2d array
The operator acting on two sites represented as d^4 x d^4 matrix.
- iiint
Apply term to the sites ii and (ii + 1).
- ll_1_termbool, optional
Boolean to indicate the term ii=L for periodic boundary conditions.
- _apply_op2_symm(Op, ii, ll_1_term, *args)[source]¶
Apply a two-site operator to the density matrix on a specified site. This is the version for density matrices with a symmetry adapted basis.
Arguments
- Opnp 2d array
The operator acting on two sites represented as d^4 x d^4 matrix.
- iiint
Apply term to the sites ii and (ii + 1).
- ll_1_termbool, optional
Boolean to indicate the term ii=L for periodic boundary conditions.
- _apply_op_nosymm(Op, idx)[source]¶
Apply an operator/gate on an arbitrary number of sites. The gate is defined on Hilbert space, not Liouville space.
Arguments
- Opnp 2d array
The operator acting on the site represented as matrix.
- idxlist of integers
The sited to be contracted with the operator.
- _apply_op_symm(Op, idx)[source]¶
Apply an operator/gate on an arbitrary number of sites to a symmetry conserving density matrix. The gate is defined on Hilbert space, not Liouville space.
Arguments
- Opnp 2d array
The operator acting on the site represented as matrix.
- idxlist of integers
The sites to be contracted with the operator.
- _dot_hmpo_mat_nosymm(Hmpo, Operators, param, quenchtime, maxdim)[source]¶
Multiply a MPO which can represent the Hamiltonian or a measurement with the density matrix. For arguments look into documention of
exactdiag.DensityMatrix.dot_hmpo_mat(). This is the version for density matrices without symmetry adapted basis. Intended for internal use.
- _dot_hmpo_mat_symm(Hmpo, Operators, param, quenchtime, maxdim)[source]¶
Multiply a MPO which can represent the Hamiltonian or a measurement with the density matrix. For arguments look into documention of
exactdiag.DensityMatrix.dot_hmpo_mat(). This is the version for density matrices with symmetry adapted basis.
- _dot_hmpo_vec_nosymm(Hmpo, Operators, param, quenchtime, maxdim)[source]¶
Multiply a MPO which represents the Liouville operator with the density matrix represented as superket vector. For arguments look into the documentation of
exactdiag.DensityMatrix.dot_hmpo_vec(). This is the version for density matrices without symmetries. Intended for internal use.
- _dot_hmpo_vec_symm(Hmpo, Operators, param, quenchtime, maxdim)[source]¶
Multiply a MPO which represents the Liouville operator with the density matrix represented as superket vector. For arguments look into the documentation of
exactdiag.DensityMatrix.dot_hmpo_vec(). This is the version for density matrices with symmetry adapted basis. Intended for internal use.
- _fidelity_with_state(Phi)[source]¶
Return the fidelity between the density matrix and a quantum state. This is possible independent of the size of the density matrix, since the fidelity simplifies to
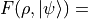
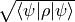 .
.Arguments
- Phiinstance of
exactdiag.PureStateor numpy 1D array Calculate the fidelity to this state vector.
- Phiinstance of
- _matrix_exp_trotter(Liou)[source]¶
Taking the matrix exponential of the Liouville operator to obtain the propagator. Without symmetries, the propagator is returned as matrix. With symmetries, a dictionary is returned where the key is the row index and the value are the four local indices plus the matrix element. In the latter case the blockdiagonal structure is used.
Arguments
- Lioumatrix
a matrix representing the Liouville operator on two sites in the Liouville space.
Details
Functions accesses class variabes
self.bidx, a dictionary of tuples orNone. IfNone, no symmetries are used. If symmetries are used, bidx is a dictionary where each set of tuples contains the indices for the same symmetry sector and the corresponding site indices.
- _traceout_first_nosymm()[source]¶
Return the reduced density matrix tracing over the first site in the system. This functions treats density matrices without symmetry.
- _traceout_first_symm(SubSec)[source]¶
Return the reduced density matrix tracing over the first site in the system. This function treats density matrices with symmetry and calculates the new density matrix in a symmetry adapted space.
Arguments
- SubSecinstance of
exactdiag.SymmetrySector This is the symmetry sector of the system from site 2 to site L (end of the system). It is calculated before hand in order to estimate if a symmetry adapted basis is appropriate.
- SubSecinstance of
- _traceout_first_symm2nosymm(SubSec)[source]¶
Return the reduced density matrix tracing over the first site in the system. This function treats density matrices with symmetry and calculates the new density matrix in the complete Hilbert space.
Arguments
- SubSecinstance of
exactdiag.SymmetrySector This is the symmetry sector of the system from site 2 to site L (end of the system). It is calculated before hand in order to estimate if a symmetry adapted basis is appropriate.
- SubSecinstance of
- _traceout_last_nosymm()[source]¶
Return the reduced density matrix tracing over the last site in the system. This functions treats density matrices without symmetry.
- _traceout_last_symm(SubSec)[source]¶
Return the reduced density matrix tracing over the last site in the system. This function treats density matrices with symmetry and calculates the new density matrix in a symmetry adapted space.
Arguments
- SubSecinstance of
exactdiag.SymmetrySector This is the symmetry sector of the system from site 1 to site (L - 1) (L is the end of the system). It is calculated before hand in order to estimate if a symmetry adapted basis is appropriate.
- SubSecinstance of
- _traceout_last_symm2nosymm(SubSec)[source]¶
Return the reduced density matrix tracing over the last site in the system. This function treats density matrices with symmetry and calculates the new density matrix in the complete Hilbert space.
Arguments
- SubSecinstance of
exactdiag.SymmetrySector This is the symmetry sector of the system from site 1 to site (L - 1) (L is the end of the system). It is calculated before hand in order to estimate if a symmetry adapted basis is appropriate.
- SubSecinstance of
- _traceout_nosymm(trce, keep)[source]¶
Trace over a set of sites specified via their indices for states without a symmetry.
Arguments
- trcelist of ints
Trace out over the indices contain in this list.
- keeplist of ints
Keep the sites whose indices are listed here. Together trce and keep should list all indices exactly one time.
- _traceout_symm(trce, keep, SubSec=None)[source]¶
Trace over a set of sites specified via their indices for states with symmetry returning a density matrix considering the symmetry in the larger Hilbert space.
Arguments
- trcelist of ints
Trace out over the indices contain in this list.
- keeplist of ints
Keep the sites whose indices are listed here. Together trce and keep should list all indices exactly one time.
- SubSecinstance of
exactdiag.SymmetrySector If symmetry sector is already calculated, it can be passed with this argument. Otherwise pass
None. Default toNone.
- _traceout_symm2nosymm(trce, keep, SubSec=None)[source]¶
Trace over a set of sites specified via their indices for states with symmetry returning a density matrix not considering the symmetry.
Arguments
- trcelist of ints
Trace out over the indices contain in this list.
- keeplist of ints
Keep the sites whose indices are listed here. Together trce and keep should list all indices exactly one time.
Details
This functions is unefficient when tracing over single sites (or small subsystems).
- _trotter_finalize(*args)[source]¶
Density matrices without symmetry-adapted basis are converted from the rank-2L tensor back to their matrix representation.
- _trotter_initialize(qt)[source]¶
Density matrices without symmetry-adapted basis are converted to a rank-2L tensor.
Arguments
- qtboolean
If
True, an error is raised because quantum trajectories on a density matrix are not supported. IfFalse, then ok.
- distance(State, dist)[source]¶
Measure the distance between the pure state and another quantum state.
Arguments
- State1d or 2d numpy array
Represents another pure state (1d) or a density matrix (2d).
- distancestr
Define the type of distance measure. Either
TraceorInfidelity.
- dot_hmpo_mat(Hmpo, Operators, param, quenchtime, maxdim)[source]¶
Multiply a Hamiltonian represented as MPO / MPO rule set with a density matrix.
Arguments
- Hmpoinstance of
MPO.MPO Hamiltonian represented in the MPO class or MPO representing a measurement.
- Operatorsinstance of
Ops.OperatorList(or dictionary) Operators defined used in the simulation.
- paramdictionary
dictionary representing a MPS simulation.
- quenchtimetuple of int and float
Storing the number of the quench and the actual time
- maxdimint
maximal dimension of the Hilbert space. Only referenced in …
- Hmpoinstance of
- dot_hmpo_vec(Hmpo, Operators, param, quenchtime, maxdim)[source]¶
Multiply a Liouville operator in the Liouville space represented as MPO / MPO rule set with a density matrix represented as superket. The field
self.rhohas to be converted to the superket before calling the subroutine. Result is returned as superket as well.Arguments
- Hmpoinstance of
MPO.MPO Hamiltonian represented in the MPO class or MPO representing a measurement.
- Operatorsinstance of
Ops.OperatorList(or dictionary) Operators defined used in the simulation.
- paramdictionary
dictionary representing a MPS simulation.
- quenchtimetuple of int and float
Storing the number of the quench and the actual time
- maxdimint
maximal dimension of the Hilbert space. Only referenced in …
- Hmpoinstance of
- fidelity(Phi)[source]¶
Return the fidelity. Not implemented for any density matrix yet.
Arguments
- Phipure state or density matrix
Calculate fidelity to this state. It can be represented as 1d or 2d numpy array, an instance of
exactdiag.PureState, or as instance ofexactdiag.DensityMatrix.
Details
Implementation for density matrices is missing to avoid calculating the square root of large matrices.
- get_bond_entropies()[source]¶
Function to return dummy value for bond entropy. There is no bond entropy defined right now since the eigenvalues do not hold the relation any more.
Details
We could split via SVD for each bipartition.
- meas_mpo(Hmpo, Operators, param, quenchtime, maxdim)[source]¶
Measure a MPO using the rule set.
Arguments
- Hmpoinstance of
MPO.MPO Hamiltonian represented in the MPO class or MPO representing a measurement.
- Operatorsinstance of
Ops.OperatorList(or dictionary) Operators defined used in the simulation.
- paramdictionary
dictionary representing a MPS simulation.
- quenchtimetuple of int and float
Storing the number of the quench and the actual time
- maxdimint
maximal dimension of the Hilbert space. Only referenced in …
- Hmpoinstance of
- propagate_expm(dt, Hmpo, param, sparse, maxdim, quenchtime, qt)[source]¶
Propagate a state in the Liouville space according to the Lindblad master equation.
Arguments
- dtfloat
time step for evolution.
- Hmpoinstance of
MPO.MPO Hamiltonian represented in the MPO class or MPO representing a measurement.
- paramdictionary
dictionary representing a MPS simulation.
- sparseboolean
flag if sparse matrix should be used for building Hamiltonian.
- maxdimint
maximal dimension of the Hilbert space. Only referenced in …
- quenchtimetuple of int and float
Storing the number of the quench and the actual time
- qtboolean
If
True, evolution should be carried out with quantum trajectories which is impossible for density matrices and therefore an exception is raised. IfFalse, evolution in Liouville space is ok.
- propagate_gates(*args, **kwargs)[source]¶
Propagation with gates is not implemented for density matrices
- propagate_krylov(dt, Hmpo, Operators, param, sparse, maxdim, quenchtime, qt)[source]¶
Propagation with Krylov using the Arnoldi algorithm.
Arguments
- dtfloat
time step for evolution.
- Hmpoinstance of
MPO.MPO Hamiltonian represented in the MPO class or MPO representing a measurement.
- Operatorsinstance of
Ops.OperatorList contains the operators defined on the local Hilbert space.
- paramdictionary
dictionary representing a MPS simulation.
- sparseboolean
Build Liouville operators as sparse or dense matrix. Only accessed in edlibmode=1.
- maxdimint
maximal dimension of the Hilbert space. Only referenced in …
- quenchtimetuple of int and float
Storing the number of the quench and the actual time
- qtboolean
If
True, evolution should be carried out with quantum trajectories which is impossible for density matrices and therefore an exception is raised. IfFalse, evolution in Liouville space is ok.
Details
The following modes are available for the Krylov time evolution, which is set in the convergence criteria for the quenches, i.e.,
convergence.KrylovConvParamwith the variableedlibmode:0 : Built Hamiltonian as matrix and use built-in scipy method.
1 : Built Hamiltonian as matrix and use EDLib Krylov.
2 : Use dot product of MPO and state vector and EDLib Krylov.
3 : As 2, but save Krylov vectors to hard disk to reduce RAM memory usage.
-1 : default. For :math`dim < 2^{10}` take edlibmode=0, then for
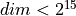 use edlibmode=2, otherwise edlibmode=3
use edlibmode=2, otherwise edlibmode=3
Since the Liouville operator is in general not hermitian, Krylov uses the Arnoldi implementation of the algorithm.
- trotter_layer(dt, Hmpo, Operators, param, quenchtime, even, *args)[source]¶
Apply two site operators for the Trotter decomposition for the odd or even layer.
Arguments
- dtfloat
time step for evolution.
- Hmpoinstance of
MPO.MPO Hamiltonian represented in the MPO class or MPO representing a measurement.
- Operatorsinstance of
Ops.OperatorList contains the operators for building the Hamiltonian and observables.
- paramdictionary
dictionary representing a MPS simulation.
- quenchtimetuple of int and float
Storing the number of the quench and the actual time
- evenboolean
If
True, even layer of operators is applied on sites (2k , 2k + 1). IfFalse, we use the odd layer.
- exactdiag.idx_symmetry_blocks(Operators, param, ll=2)[source]¶
Generates the indices for blocks conserving the symmetry.
Arguments
- Operatorsinstance of
Ops.OperatorList contains all operators, amongst them the diagonal generators of the symmetry.
- paramdictionary
containing the string identifiers for the generators of the U(1) and Z2 symmetries.
- llint, optional
Number of sites to generate the blocks Default to 2.
Details
If no symmetries are present, the funtion returns
None. Otherwise the following output in form of a dictionary is generated. The keys of the dictionary are a tuple of indices in the combined Hilbert space of ll sites with the same quantum number. The value for the corresponding key is a tupled tuple. The i-th tuple contains the local indices for the i-th combined index.- Operatorsinstance of
- exactdiag.Lidx_symmetry_blocks(Operators, param, ld, ll=2)[source]¶
Function analogous to
idx_symmetry_block, but constructing the mapping for the Liouville space used for density matrices:Arguments
- Operatorsinstance of
Ops.OperatorList contains all operators, amongst them the diagonal generators of the symmetry.
- paramdictionary
containing the string identifiers for the generators of the U(1) and Z2 symmetries.
- ldinteger
local dimension of the Hilbert space.
- llint, optional
Number of sites to generate the blocks Default to 2.
- Operatorsinstance of